svCapture
The svCapture pipeline and app combine two functionalities - error-correction and SV junction identification - optimized for detecting rare structural variants (SVs) in short-read, whole-genome sequencing libraries subjected to target capture, typically by probe hybridization.
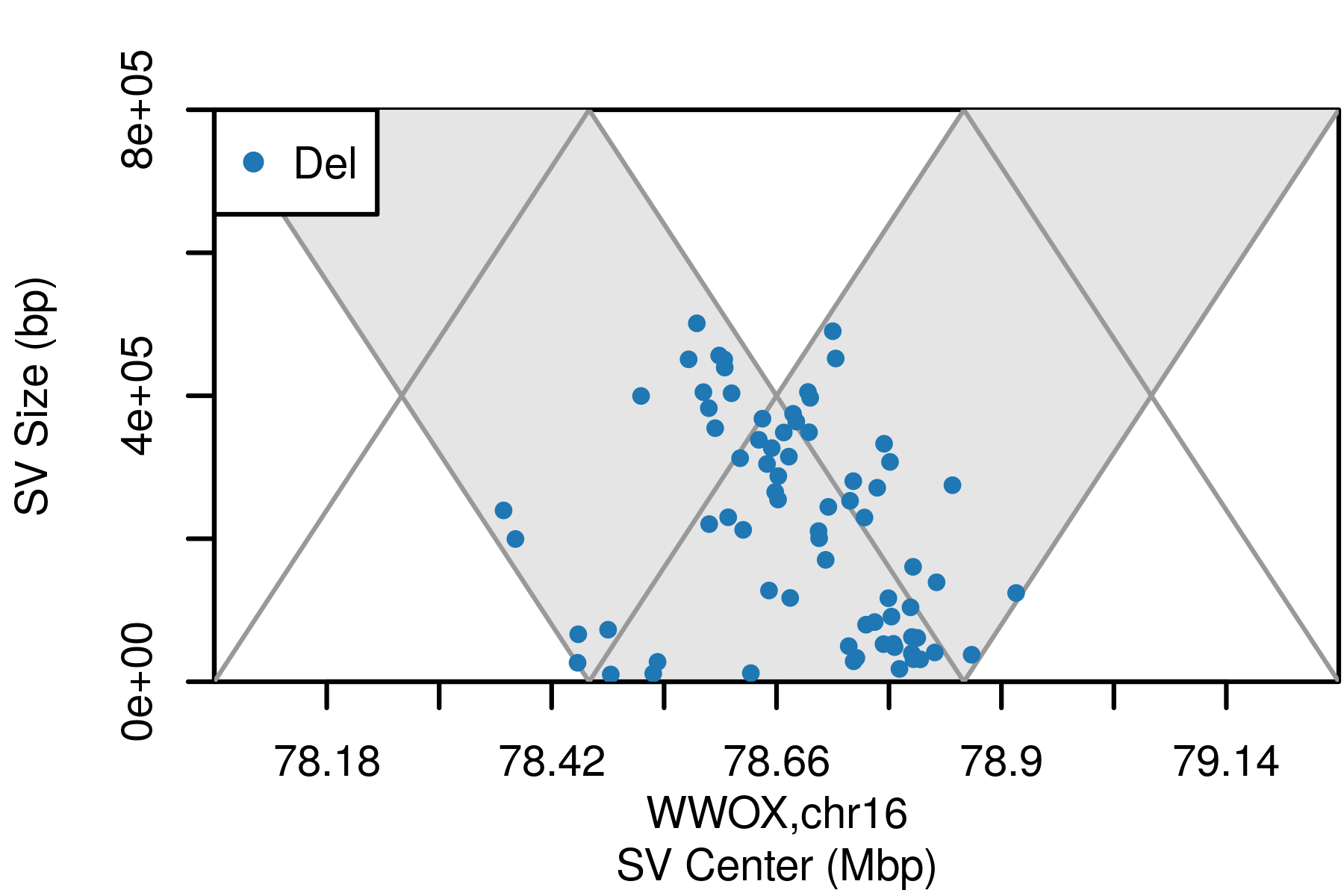
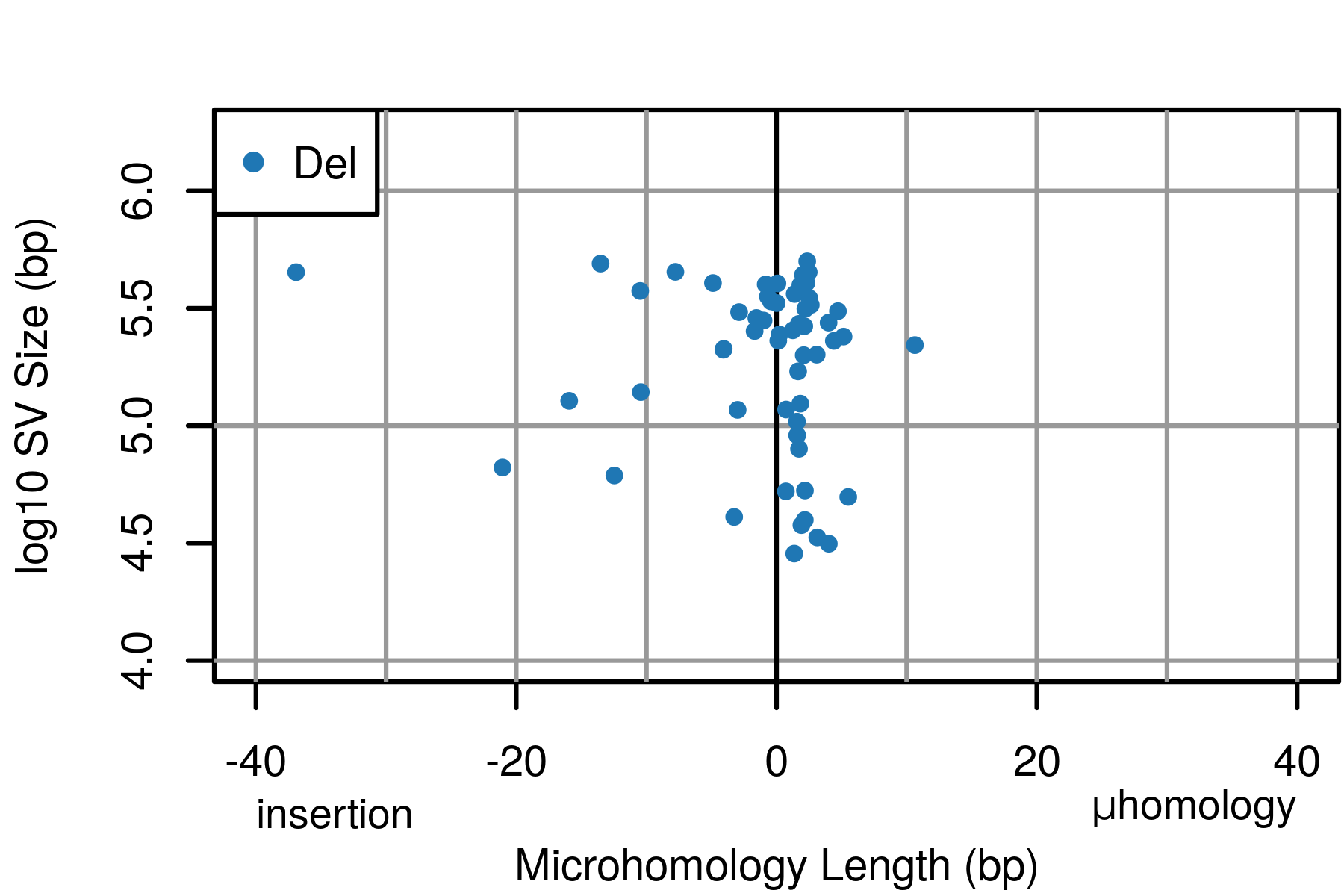
The motivation for svCapture was to extend the principles of error-corrected duplex sequencing to SVs, beyond SNVs/indels. Doing so requires careful attention to the distinct error modes by which false SVs and SNVs arise, and simultaneous bookkeeping of both source molecule DNA strands and alignment discontinuities. Once satisfied, confident detection of even single-molecule, nonhomologous SVs can be asserted.
Citation
svCapture and its use for SV identification is described in detail here:
Job scripts, log files and resource files associated with the work in this publication are available here (a simpler demo is available via the link at the left):